The principle of minimal frustration does not rule out that some energetic frustration may be present in a folded protein. Moreover, the remaining frustration may facilitate motion of the protein around its native basin, and as such the residual frustration may be fundamental to protein function [9, 10].
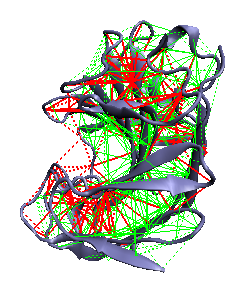
Theoretical methods allow for spatially localizing and quantifying the energetic frustration present in native protein structures by developing a spatially local version of the global gap criterion formulation of the minimal frustration principle [11]. It compares the contribution to the extra stabilization energy ascribed to a given pair of amino acids in the native protein to the statistics of the energies that would be found by placing different residues in the same native location or by creating a different environment for the interacting pair. If there is a sufficient additional stabilization for an individual native pair as normalized by the typical energy fluctuation (in accord with the global Z-score criterion for minimal frustration) the local interaction can be called "minimally frustrated". If the stabilization of the native pair lies in the middle of the distribution of alternatives, the interaction can be considered "neutral". On the other hand, if the native pair is sufficiently destabilizing compared to the other possibilities the interaction is "highly frustrated". Details of the method and the energy functions can be found here [12].
How the algorithm works
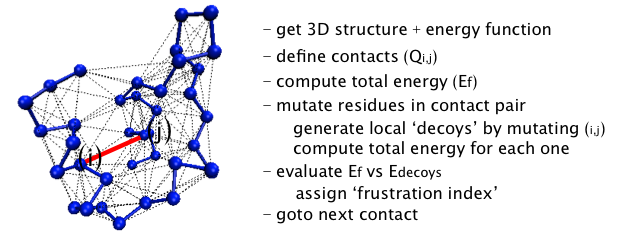
To evaluate how frustrated the interactions are in a given structure,
the protein sequence is systematically perturbed and the resulting
total energy change is computed, according to the AMW energy function
[13]. The amino acids forming a particular contact
pair are changed to other amino acids generating a set of decoys for
which the total energy of the protein is recomputed. Sequence space is
randomly sampled according to the native amino acid frequency
distribution of the particular protein under consideration, giving one
thousand appropriately distributed decoys for each contact. A
histogram of the energy of the decoys is compared to the distribution
to the native energy, HN. The "frustration index" (Fi,j) for the contact
between the amino acids i,j is defined as a Z-score of the energy of
the native pair compared to the N decoys:
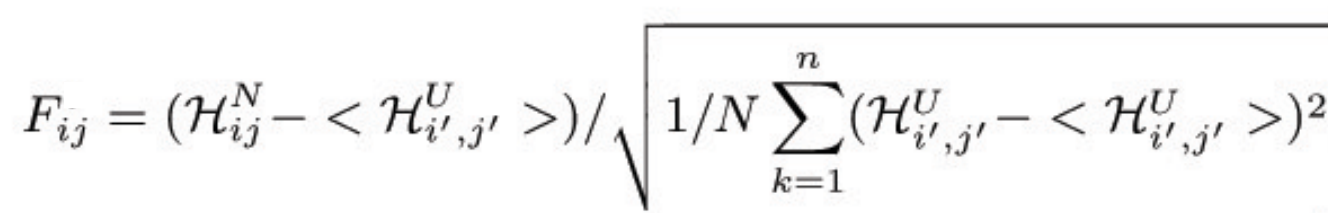
where HUi',j' is the energy of the decoy. The frustration index
measures of how favorable a particular interaction is relative to the
set of all possible interactions in that location, normalized using
the variance of that distribution.